SPANDx workflow for analysis of haploid next-generation re


Comparative genomics and antimicrobial resistance profiling of Elizabethkingia isolates reveals nosocomial transmission and in vitro susceptibility to fluoroquinolones, tetracyclines and trimethoprim-sulfamethoxazole

PDF) Emergent biomarker derived from next-generation sequencing to identify pain patients requiring uncommonly high opioid doses

Comparative genomics and antimicrobial resistance profiling of Elizabethkingia isolates reveals nosocomial transmission and in vitro susceptibility to fluoroquinolones, tetracyclines and trimethoprim-sulfamethoxazole

Figure 2 from The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data.

PDF) SPANDx: A genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets

Save time with our One-Day-Workflow - GenDx
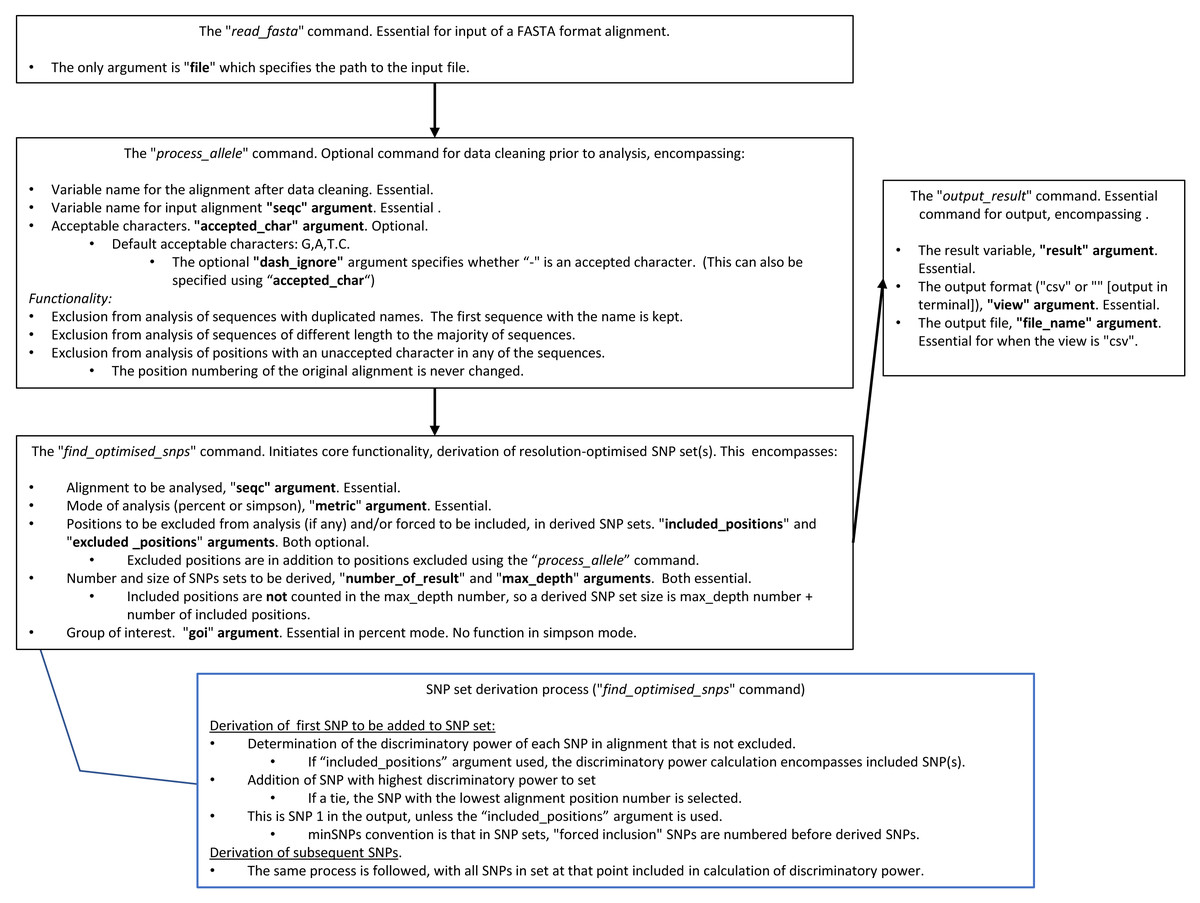
minSNPs: an R package for the derivation of resolution-optimised SNP sets from microbial genomic data [PeerJ]

Standardized phylogenetic and molecular evolutionary analysis applied to species across the microbial tree of life. - Abstract - Europe PMC

SPANDx workflow for analysis of haploid next-generation re-sequencing data.

Simultaneous identification of Haemophilus influenzae and Haemophilus haemolyticus using real-time PCR

Derek SAROVICH, Senior Research Fellow, PhD Microbiology, University of the Sunshine Coast, USC, Faculty of Science, Health, Education and Engineering

Metabolism of ʟ -arabinose converges with virulence regulation to promote enteric pathogen fitness
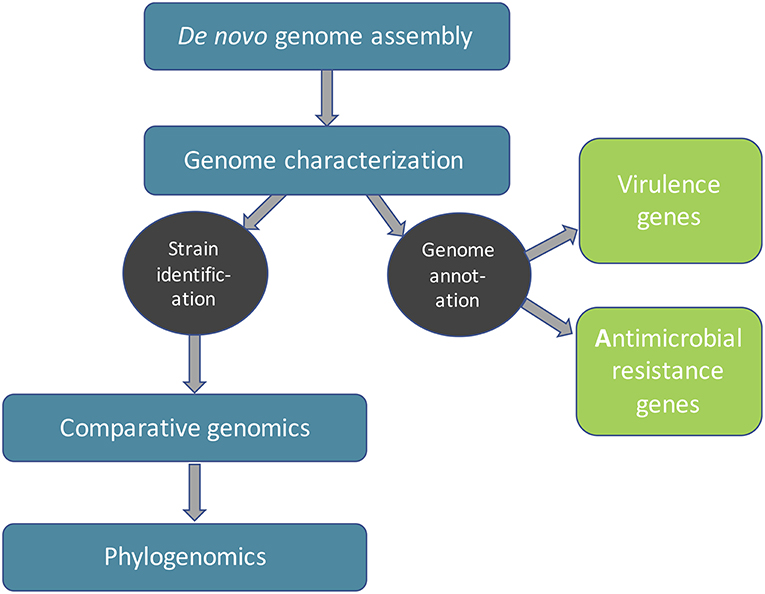
Frontiers Machine Learning Approaches for Epidemiological Investigations of Food-Borne Disease Outbreaks

SPANXB1 Gene - GeneCards, SPNXB Protein